Distance sampling analysis in unmarked
Richard Chandler, USGS Patuxent Wildlife Research Center
March 4, 2020
Source:vignettes/distsamp.Rmd
distsamp.RmdAbstract
Distance sampling is a wildlife sampling technique used to estimate
population size or density. Describing how density varies spatially is
often of equal interest; however, conventional methods of analysis do
not allow for explicit modeling of both density and detection
probability. The function distsamp implements the
multinomial-Poisson mixture model of Royle et al.
(2004), which was developed to overcome this limitation. This
model requires that line- or point-transects are spatially replicated
and that distance data are recorded in discrete intervals. The function
extends this basic model, by introducing the parameter
,
the probability of being available for detection (Chandler et al. 2011). Furthermore, this
function allows abundance to be modeled using the negative binomial
distribution, which may be useful for dealing with over-dispersion. This
document describes how to format data, fit models, and manipulate
results in package . It does not cover the statistical theory and
assumptions underlying distance sampling (Buckland et al. 2001), which the user is
expected to be familiar with.
Introduction
Spatial variation in density is common to virtually all wildlife populations, and describing this variation is a central objective of much research. To accurately estimate and model density, it is often necessary to account for individuals present but not detected. Distance from observer is a ubiquitous source of variation in detection probability, and thus distance sampling has become a commonly used survey methodology. Numerous options exist for analyzing distance sampling data, but here the focus is on the model of Royle et al. (2004), which assumes that multiple transects have been surveyed and distance data are recorded in discrete intervals. The details of the model formulation are as follows:
The latent transect-level abundance distribution is currently assumed to be The detection process is modeled as where is the multinomial cell probability for transect in distance class . These are computed by integrating a detection function such as the half-normal (with scale parameter ) over each distance interval.
Parameters and can be vectors affected by transect-specific covariates using the log link.
Importing, formatting, and summarizing data
The first step is to import the data into R. The simplest option is
to use the read.csv function to import a .csv file that has
been formatted so that each row represents a transect, and columns
describe either the number of individuals detected in each distance
interval or transect-specific covariates. Alternatively, if data were
not recorded in discrete distance intervals, a .csv file could be
imported that contains a row for each individual detected and columns
for the distances and transect names. This could then be converted to
transect-level data using the function formatDistData. For
example,
library(unmarked)
dists <- read.csv(system.file("csv", "distdata.csv", package="unmarked"),
stringsAsFactors=TRUE)
head(dists, 10)## distance transect
## 1 1 a
## 2 18 a
## 3 7 a
## 4 2 a
## 5 13 b
## 6 3 b
## 7 5 b
## 8 10 b
## 9 6 c
## 10 9 c## [1] "a" "b" "c" "d" "e" "f" "g"
yDat <- formatDistData(dists, distCol="distance",
transectNameCol="transect", dist.breaks=c(0, 5, 10, 15, 20))
yDat## [0,5] (5,10] (10,15] (15,20]
## a 2 1 0 1
## b 2 1 1 0
## c 2 2 0 0
## d 2 1 1 0
## e 1 0 1 2
## f 2 1 1 0
## g 0 0 0 0Here we have created an object called yDat that contains
counts for each transect (row) in each distance interval (columns). Note
the method used to include transect "g", which was surveyed
but where no individuals were detected. It is important that all
surveyed transects are included in the analysis.
Suppose there also exists transect-specific covariate data.
(covs <- data.frame(canopyHt = c(5, 8, 3, 2, 4, 7, 5),
habitat = c('A','A','A','A','B','B','B'), row.names=letters[1:7]))## canopyHt habitat
## a 5 A
## b 8 A
## c 3 A
## d 2 A
## e 4 B
## f 7 B
## g 5 BThe function unmarkedFrameDS can now be used to organize
these data along with their metadata (study design (line- or
point-transect), distance class break points, transect lengths, and
units of measurement) into an object to be used as the data
argument in distsamp. By organizing the data this way, the
user does not need to repetitively specify these arguments during each
call to distsamp, thereby reducing the potential for errors
and facilitating data summary and manipulation.
umf <- unmarkedFrameDS(y=as.matrix(yDat), siteCovs=covs, survey="line",
dist.breaks=c(0, 5, 10, 15, 20), tlength=rep(100, 7),
unitsIn="m")## Warning: siteCovs contains characters. Converting them to factors.Note that there is no obsCovs argument, meaning that
distance-interval-level covariates cannot be included in the analysis.
The call to unmarkedFrameDS indicates that the data were
collected on seven line transects, each 100 meters long, and detections
were tabulated into distance intervals defined by the
dist.breaks cutpoints. It is important that both transect
lengths and distance break points are provided in the same units
specified by unitsIn.
We can look at these data using a variety of methods.
summary(umf)## unmarkedFrameDS Object
##
## line-transect survey design
## Distance class cutpoints (m): 0 5 10 15 20
##
## 7 sites
## Maximum number of distance classes per site: 4
## Mean number of distance classes per site: 4
## Sites with at least one detection: 6
##
## Tabulation of y observations:
## 0 1 2
## 11 10 7
##
## Site-level covariates:
## canopyHt habitat
## Min. :2.000 A:4
## 1st Qu.:3.500 B:3
## Median :5.000
## Mean :4.857
## 3rd Qu.:6.000
## Max. :8.000
hist(umf, xlab="distance (m)", main="", cex.lab=0.8, cex.axis=0.8)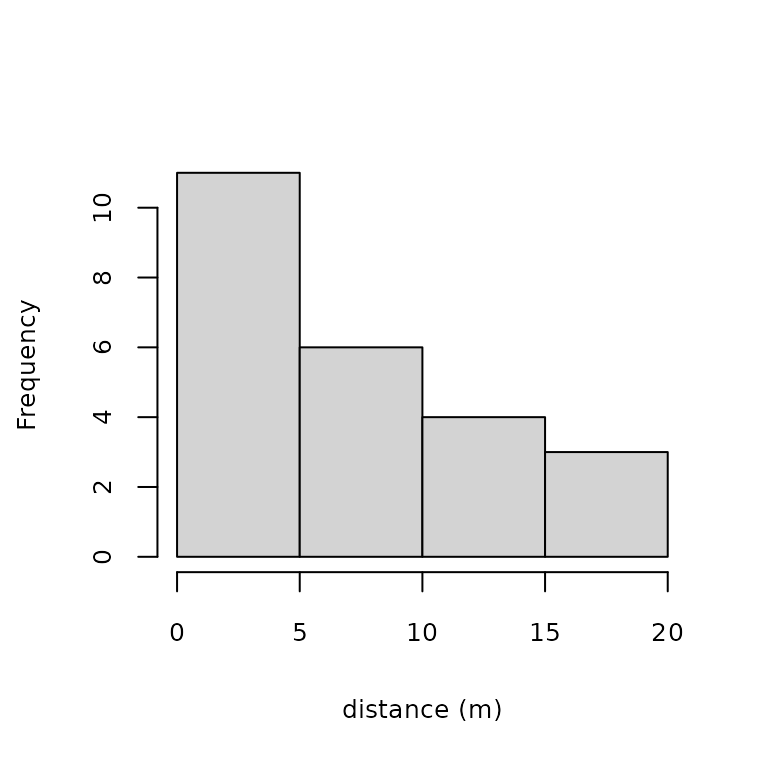
Figure 1. Histogram of detection distances
Model fitting
Now that we have put our data into an object of class
unmarkedFrameDS, we are ready to fit some models with
distsamp. The first argument is a formula
which specifies the detection covariates followed by the density (or
abundance) covariates. The only other required argument is the
data, but several other optional arguments exist. By
default, the half-normal detection function is used to model density in
animals / ha. The detection function can be selected using the
keyfun argument. The response can be changed from
"density", to "abund" with the
output argument. When modeling density, the output units
can be changed from "ha" to "kmsq" using the
unitsOut argument. distsamp also includes the
arguments starts, method, and
control, which are common to all unmarked fitting
functions.
Below is a series of models that demonstrates distsamp’s
arguments and defaults.
hn_Null <- distsamp(~1~1, umf)
hn_Null <- distsamp(~1~1, umf, keyfun="halfnorm", output="density",
unitsOut="ha")
haz_Null <- distsamp(~1~1, umf, keyfun="hazard")
hn_Hab.Ht <- distsamp(~canopyHt ~habitat, umf)The first two models are the same, a null half-normal detection function with density returned in animals / ha (on the log-scale). The third model uses the hazard-rate detection function, and the fourth model includes covariates affecting the Poisson mean () and the half-normal scale parameter ().
Once a model has been fit, typing its name will display parameter estimate information and AIC. A summary method shows extra details including the scale on which parameters were estimated and convergence results.
haz_Null##
## Call:
## distsamp(formula = ~1 ~ 1, data = umf, keyfun = "hazard")
##
## Density (log-scale):
## Estimate SE z P(>|z|)
## 2.97 0.959 3.09 0.00198
##
## Detection (log-scale):
## Estimate SE z P(>|z|)
## 1.44 2.37 0.606 0.544
##
## Hazard-rate(scale) (log-scale):
## Estimate SE z P(>|z|)
## -0.0223 1.21 -0.0185 0.985
##
## AIC: 65.17091
## Number of sites: 7
##
## Survey design: line-transect
## Detection function: hazard
## UnitsIn: m
## UnitsOut: haManipulating results
Use predict to get estimates and 95% confidence
intervals from the model on the original scale. Note that since this is
the null model, all sites will have the same estimates and we can just
look at the first row of the output.
names(haz_Null)## [1] "state" "det" "scale"
predict(haz_Null, type="state")[1,]## Predicted SE lower upper
## 1 19.39481 18.5935 2.962454 126.9753
predict(haz_Null, type="det")[1,]## Predicted SE lower upper
## 1 4.210818 9.985581 0.04034758 439.456
predict(haz_Null, type="scale")[1,]## Predicted SE lower upper
## 1 0.977985 1.179853 0.09192469 10.40476The first call to predict returns population density,
since this is the default state parameter modeled when
distsamp’s output argument is set to
"density". The second returns the hazard-rate shape
parameter, and third is the hazard-rate scale parameter.
When covariates are present, predict in conjunction with
the newdata argument should be used. Here, we request the
value of sigma when canopy height was 5 meters tall.
nd <- data.frame(canopyHt = 5)
predict(hn_Hab.Ht, type = 'det', newdata=nd, appendData = TRUE)## Predicted SE lower upper canopyHt
## 1 10.32811 2.54458 6.372452 16.73921 5Parameters that do not occur in the likelihood may also be of interest. For example, the number of individuals occurring in the sampled plots (local population size) is a fundamental parameter in monitoring and conservation efforts. The following commands can be used to derive this parameter from our model of density:
site.level.density <- predict(hn_Hab.Ht, type="state")$Predicted
plotArea.inHectares <- 100 * 40 / 10000
site.level.abundance <- site.level.density * plotArea.inHectares
(N.hat <- sum(site.level.abundance))## [1] 39.06128To describe the uncertainty of N.hat, or any other
derived parameter, we can use a parametric bootstrap approach. First we
define a function to estimate N.hat, and then we apply this
function to numerous models fit to data simulated from our original
model.
getN.hat <- function(fit) {
d <- predict(fit, type="state")$Predicted
a <- d * (100 * 40 / 10000)
N.hat <- c(N.hat = sum(a))
return(N.hat)
}
pb <- parboot(hn_Hab.Ht, statistic=getN.hat, nsim=25)
pb##
## Call: parboot(object = hn_Hab.Ht, statistic = getN.hat, nsim = 25)
##
## Parametric Bootstrap Statistics:
## t0 mean(t0 - t_B) StdDev(t0 - t_B) Pr(t_B > t0)
## N.hat 39.1 -3.79 8.64 0.692
##
## t_B quantiles:
## 0% 2.5% 25% 50% 75% 97.5% 100%
## [1,] 21 27 38 42 50 58 58
##
## t0 = Original statistic computed from data
## t_B = Vector of bootstrap samplesHere, t_B is an approximation of the sampling
distribution for N.hat, conditioned on our fitted model.
Confidence intervals can be calculated from the quantiles of
t_B. Note that in practice nsim should be set
to a much larger value and a goodness-of-fit test should be performed
before making inference from a fitted model. Parametric bootstrapping
can be used for the latter by supplying a fit statistic such as
SSE instead of getN.hat. See
?parboot and vignette('unmarked') for
examples.
Prediction and plotting
A predict method exits for all unmarkedFit
objects, which is useful when multiple covariate combinations exist.
This method also facilitates plotting. Suppose we wanted model
predictions from the covariate model along the range of covariate values
studied. First we need to make new data.frames holding the
desired covariate combinations. Note that column names must match those
of the original data, and factor variables must contain the same
levels.
head(habConstant <- data.frame(canopyHt = seq(2, 8, length=20),
habitat=factor("A", levels=c("A", "B"))))## canopyHt habitat
## 1 2.000000 A
## 2 2.315789 A
## 3 2.631579 A
## 4 2.947368 A
## 5 3.263158 A
## 6 3.578947 A
(htConstant <- data.frame(canopyHt = 5,
habitat=factor(c("A", "B"))))## canopyHt habitat
## 1 5 A
## 2 5 BNow predict can be used to estimate density and
for each row of our new data.frames.
(Elambda <- predict(hn_Hab.Ht, type="state", newdata=htConstant,
appendData=TRUE))## Predicted SE lower upper canopyHt habitat
## 1 16.22835 5.058481 8.809465 29.89506 5 A
## 2 10.91326 4.370499 4.978158 23.92436 5 B## Predicted SE lower upper canopyHt habitat
## 1 10.76998 4.259542 4.960935 23.38117 2.000000 A
## 2 10.72259 3.968438 5.191220 22.14777 2.315789 A
## 3 10.67541 3.693948 5.418117 21.03394 2.631579 A
## 4 10.62844 3.439012 5.637007 20.03965 2.947368 A
## 5 10.58167 3.207199 5.842026 19.16659 3.263158 A
## 6 10.53511 3.002718 6.026005 18.41826 3.578947 AOnce predictions have been made, plotting is straight-forward. Figure 2a shows density as a function of habitat type, and Figure 2b shows that is not related to canopy height.
par(mfrow=c(1, 2))
with(Elambda, {
x <- barplot(Predicted, names=habitat, xlab="Habitat",
ylab="Density (animals / ha)", ylim=c(0, 25), cex.names=0.7,
cex.lab=0.7, cex.axis=0.7)
arrows(x, Predicted, x, Predicted+SE, code=3, angle=90, length=0.05)
box()
})
with(Esigma, {
plot(canopyHt, Predicted, type="l", xlab="Canopy height",
ylab=expression(paste("Half-normal scale parameter (", sigma, ")",
sep="")), ylim=c(0, 20), cex=0.7, cex.lab=0.7,
cex.axis=0.7)
lines(canopyHt, Predicted+SE, lty=2)
lines(canopyHt, Predicted-SE, lty=2)
})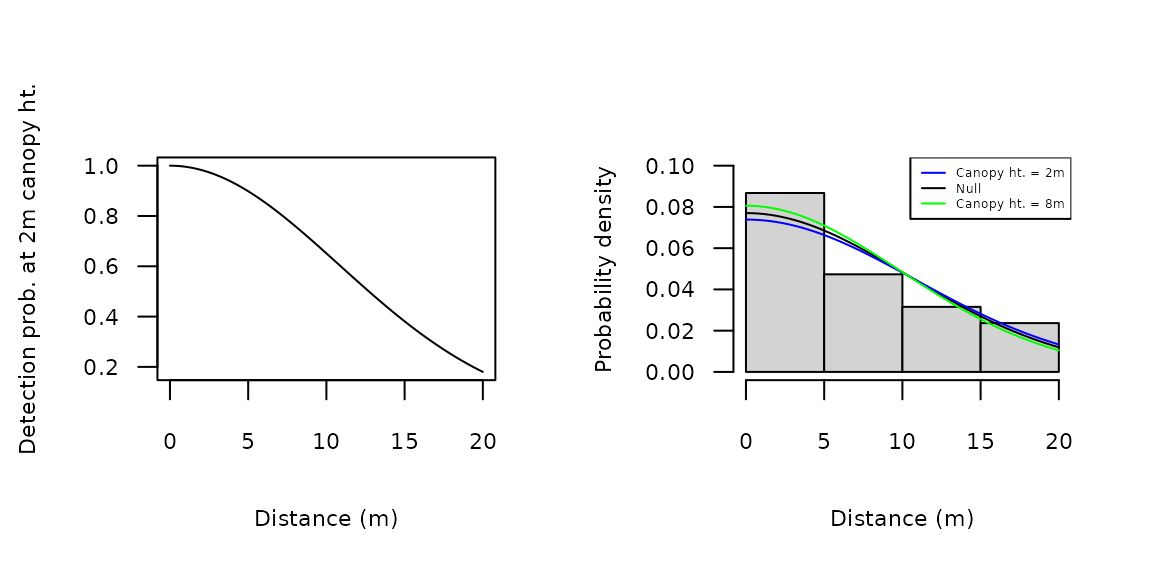
Figure 2. Predicted covariate relationships
Plots of the detection function parameters can be less informative
than plots of the detection functions themselves. To do the latter, we
can plug predicted values of
at given covariate values into the gxhn function. For
instance, Figure 3a shows to the half-normal function at a canopy height
of 2m. This was plotted by setting
to 10.8, the predicted value shown above. The available detection
functions are described on the detFuns help page.
Probability density functions such as dxhn can be plotted
with the distance histogram using the hist method for
unmarkedFitDS objects (Figure 3b). This only works for
models without detection covariates; however, probability density
functions at specific covariate values can be added in a fashion similar
to that above (Figure 3b).
par(mfrow=c(1, 2))
plot(function(x) gxhn(x, sigma=10.8), 0, 20, xlab="Distance (m)",
ylab="Detection prob. at 2m canopy ht.", cex.lab=0.7,
cex.axis=0.7, las=1)
hist(hn_Null, xlab="Distance (m)", ylab="Probability density", main="",
ylim=c(0, 0.1), cex.lab=0.7, cex.axis=0.7, las=1)
plot(function(x) dxhn(x, sigma=10.8), 0, 20, add=TRUE, col="blue")
plot(function(x) dxhn(x, sigma=9.9), 0, 20, add=TRUE, col="green")
legend('topright', c("Canopy ht. = 2m", "Null", "Canopy ht. = 8m"),
col=c("blue", "black", "green"), lty=1, cex=0.4)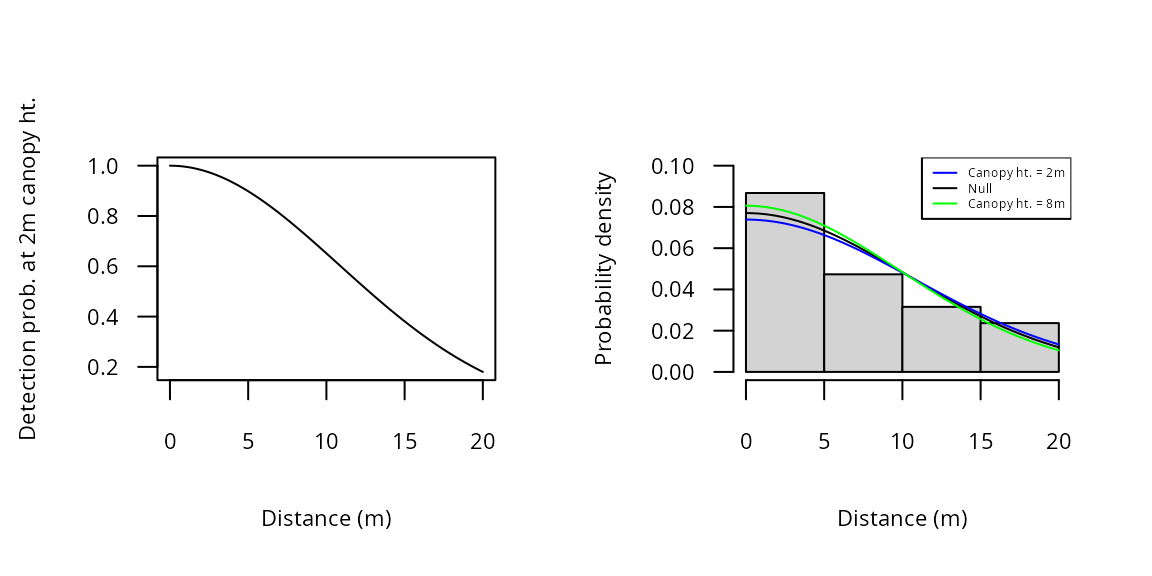
Figure 3. Detection and probability density functions
Model extensions
A common criticism of distance sampling is that all individuals must
be available for detection. Similarly, the probability of detecting an
individual a distance of 0 must be 1. These assumptions often cannot be
met. For instance, when counting cues such as bird songs or whale blows,
the probability that an individual will produce a cue (and thus be
available for detection) is rarely 1 during the sampling interval.
Recently developed methods (Chandler et al.
2011) allow for the estimation of the probability of being
available for detection
.
To do so, replicate distance sampling observations must be collected at
each transect. These replicates could be collected using repeated visits
or multiple observers working independently. Implementation of this
model in unmarked is accomplished using the
gdistsamp function. The function also provides the option
to model abundance using the negative binomial distribution. Formatting
data and specifying models is similar to methods described above and is
more fully outlined in the help pages.
Conclusion
This document has emphasized methods tailored to distance sampling
analysis; however, the more general methods available in package
unmarked can also be applied to models fitted using
distsamp and gdistsamp. For example,
model-selection and model-averaging can be accomplished using the
fitList function and the modSel and
predict methods.